
Pythonを使ったプロテオミクスデータのVolcano plot例
はじめに
コントロール細胞(WT細胞)と,ある遺伝子をノックアウトした細胞(KO細胞)に発現するタンパク質群について,増減を比較し,その増減が統計的に有意なのかを,ボルケーノプロットにより描画・検討する。
ボルケーノプロット(Volcano plot):
縦軸が群間比 (WTとKOのlog2(fold change)) ,横軸がp値 (-log10(p値) )の散布図。
データ例のDL
こちらのリンクに置いてあるcsv.ファイルを例として使用する
(ファイル名:example-of-protein_expression07082022.csv)
データの読み込み
使用するライブラリのimport
import math
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as snsファイルの置いてある場所を指定して,csvファイルをpythonで読み込み,
先頭5行のデータを確認。
df = pd.read_csv('./example-of-protein_expression07082022.csv')
df.head()
(-log10(pvalue)):p-valueに対して10が底のlogをとり,−1倍したもの
データ数を確認(今回は668種類のデータ)
df.shape
先頭行は列名
ボルケーノプロットの作成
以下のプログラムを事前に作成したのでこれを実行する
class Volcano():
def __init__(self, protein_id, ratio, pval, pcutoff, min_log2ratio, max_log2ratio):
self.df = pd.DataFrame({'ID':protein_id,'Ratio(WTvsKO)': ratio,'-log10(p-value)':pval,})
self.pcutoff = pcutoff
self.min_log2 = min_log2ratio
self.max_log2 = max_log2ratio
def get_volcano(self):
"""
about parameter:
P < 0.05: -pcutoff =0.05
-2 < log2(x): min_log2ratio = -2
log2(x) < 2: min_log2ratio = 2
"""
X = self.df['Ratio(WTvsKO)'].values
y = self.df['-log10(p-value)'].values
# pickup statistically increase (p < pcutoff(ex,0.05) , 2 < max_log2ratio(ex,2))
p_cutoff = -1* math.log(self.pcutoff,10)
df_increase = self.df[(self.df['Ratio(WTvsKO)'] >= self.max_log2) & (self.df['-log10(p-value)'] > p_cutoff)]
Xincrease = df_increase['Ratio(WTvsKO)'].values
yincrease = df_increase['-log10(p-value)'].values
gene_incraese = df_increase['ID'].values
# pickup statistically decrease (p < pcutoff(ex,0.05) , min_log2ratio < log2x (ex,2))
p_cutoff = -1 * math.log(self.pcutoff, 10)
df_decrease = self.df[(self.df['Ratio(WTvsKO)'] <= self.min_log2) & (self.df['-log10(p-value)'] > p_cutoff)]
Xdecrease = df_decrease['Ratio(WTvsKO)'].values
ydecrease = df_decrease['-log10(p-value)'].values
gene_decraese = df_decrease['ID'].values
sns.set()
sns.set_context('talk')
fig,ax = plt.subplots(figsize = (12,10))
plt.style.use('ggplot')
ax.set(xlabel =('Log2(ko/wt)'),ylabel=('-Log10(p-value)'), title=("Volcano Plot\n(p-value vs Fold change)"), xlim =(-4,8))
ax.plot(X,y, marker='o',markersize=5, color = 'gray', linewidth=0)
# Add bordar line (log2(ratio)< -2, 2<log2(ratio))
ax.axvline(x= self.min_log2,color = 'black', linewidth = 0.5)
ax.axvline(x= self.max_log2, color = 'black', linewidth = 0.5)
# Add bordar line of p = 0.05
ax.axhline(y= p_cutoff, xmin=-4, xmax=8, color = 'black', linewidth = 0.5)
# Add text, p = 0.05
ax.text(8.1 , p_cutoff-0.05, 'p =' + str(self.pcutoff), size = 20, fontstyle="italic")
# Plot increasing genes
for j in range(0,len(gene_incraese)):
ax.plot(Xincrease[j], yincrease[j], marker='o', markersize=5, color = 'red', linewidth=0)
ax.annotate(gene_incraese[j],(Xincrease[j], yincrease[j]),
xytext=(0, 10), textcoords='offset points', size = 20, color = "black")
# Plot decreasing genes
for k in range(0,len(gene_decraese)):
ax.plot(Xdecrease[k], ydecrease[k], marker='o', markersize=5, color = 'blue', linewidth=0)
ax.annotate(gene_decraese[k],(Xdecrease[k], ydecrease[k]),
xytext=(0, 10), textcoords='offset points', size = 20, color = "black")
plt.show()
return df_increase, df_decrease必要な変数:
上記3行目,"def __init__(self," 以降にある以下の項目を指定する
protein_id: df["ProteinID"]
ratio: df['log2(ratio)']
pval: df['(-log10(pvalue))']
pcutoff: p-valueの最大値(今回は0.05。つまり0.05より大きいものは除外)
min_log2ratio: -log2(ratio)の最小値(今回は-1を指定。つまり2^-1より小さい物を指定)
max_log2ratio: log2(ratio)の最小値(今回は1を指定。つまり2^1より大きいものを指定)
変数の指定
上記のプログラムを実行するために3つの変数を指定
p_cutoff = 0.05
min_log2ratio = -1
max_log2ratio = 1インスタンス作成(volcano_plot)
volcano_plot = Volcano(df['ProteinID'],df['log2(ratio)'],\
df['(-log10(pvalue))'],p_cutoff,min_log2ratio,max_log2ratio)
Volcano plotの描画(get_volcano())
描画の際,統計的有意(p<0.05)かつ,2倍以上増加したものと2倍以上減少したものが返り値として得られる。
inc,dec = volcano_plot.get_volcano()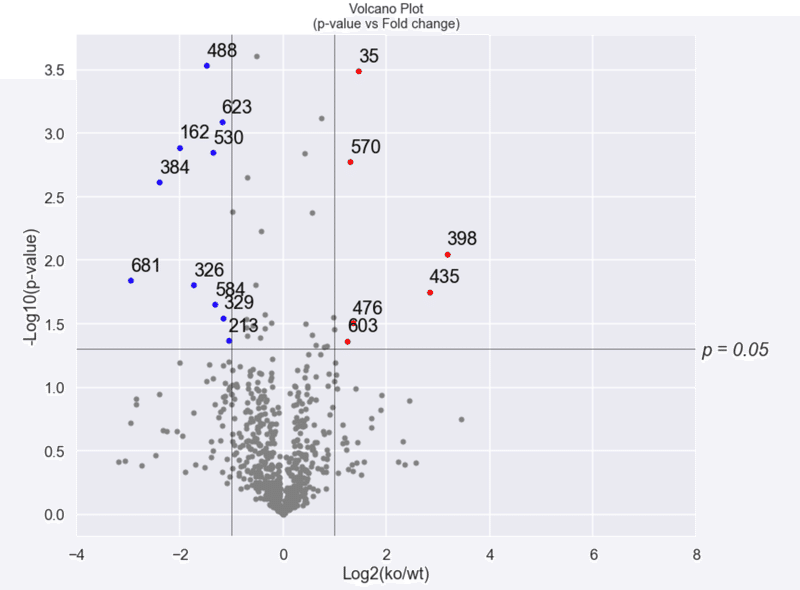
赤:2^1以上の増加かつp<0.05
増加したProteinと減少したProteinを確認
Print(inc)
print(dec)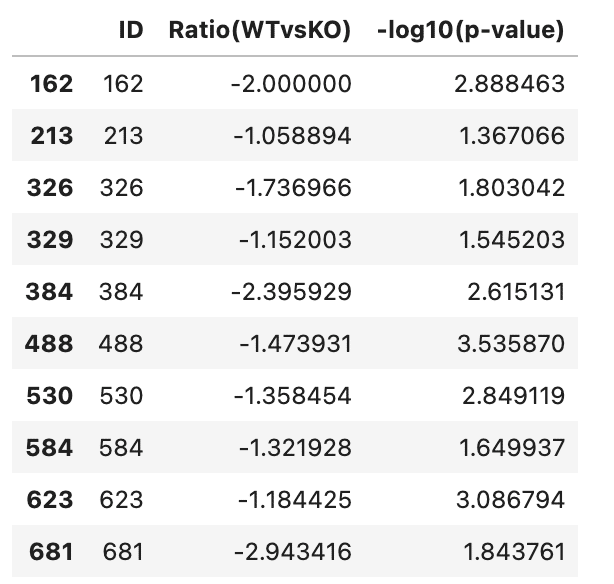
※ p_cutoff = 0.05, min_log2ratio = -1, max_log2ratio = 1 の数値を変化させることで選択範囲が変わるので,試してみると面白い
(例) p-value<0.01, -0.5≤Log2(ko/wt)≤0.5
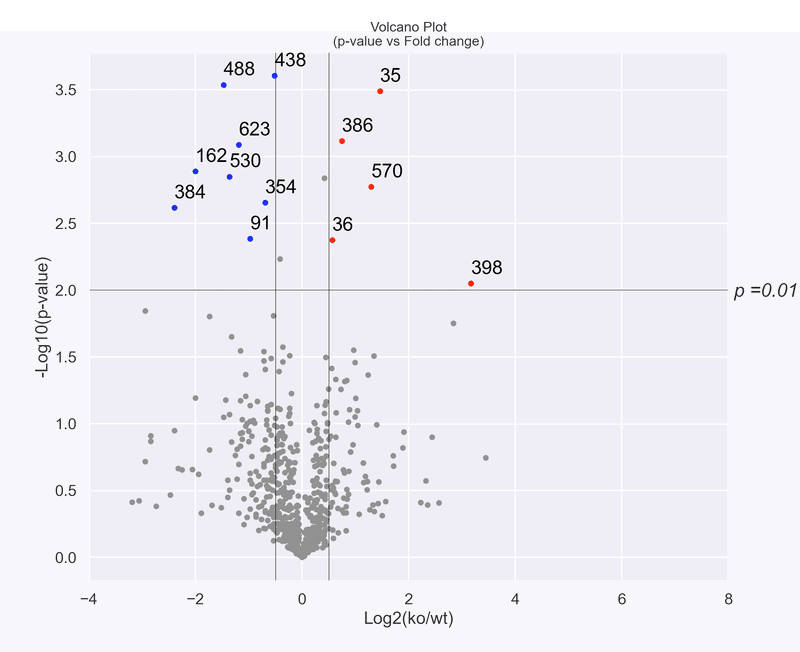
完了
おまけ:
上記実行ファイルをまとめたもの
#!/usr/bin/env python
# coding: utf-8
import math
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
#import data
df = pd.read_csv('./example-of-protein_expression07082022.csv')
df.describe()
df.head()
df.shape
# program for volcano plot
class Volcano():
def __init__(self, protein_id, ratio, pval, pcutoff, min_log2ratio, max_log2ratio):
self.df = pd.DataFrame({'ID':protein_id,'Ratio(WTvsKO)': ratio,'-log10(p-value)':pval,})
self.pcutoff = pcutoff
self.min_log2 = min_log2ratio
self.max_log2 = max_log2ratio
def get_volcano(self):
"""
about parameter:
P < 0.05: -pcutoff =0.05
-2 < log2(x): min_log2ratio = -2
log2(x) < 2: min_log2ratio = 2
"""
X = self.df['Ratio(WTvsKO)'].values
y = self.df['-log10(p-value)'].values
# pickup statistically increase (p < pcutoff(ex,0.05) , 2 < max_log2ratio(ex,2))
p_cutoff = -1* math.log(self.pcutoff,10)
df_increase = self.df[(self.df['Ratio(WTvsKO)'] >= self.max_log2) & (self.df['-log10(p-value)'] > p_cutoff)]
Xincrease = df_increase['Ratio(WTvsKO)'].values
yincrease = df_increase['-log10(p-value)'].values
gene_incraese = df_increase['ID'].values
# pickup statistically decrease (p < pcutoff(ex,0.05) , min_log2ratio < log2x (ex,2))
p_cutoff = -1 * math.log(self.pcutoff, 10)
df_decrease = self.df[(self.df['Ratio(WTvsKO)'] <= self.min_log2) & (self.df['-log10(p-value)'] > p_cutoff)]
Xdecrease = df_decrease['Ratio(WTvsKO)'].values
ydecrease = df_decrease['-log10(p-value)'].values
gene_decraese = df_decrease['ID'].values
sns.set()
sns.set_context('talk')
fig,ax = plt.subplots(figsize = (12,10))
plt.style.use('ggplot')
ax.set(xlabel =('Log2(ko/wt)'),ylabel=('-Log10(p-value)'), title=("Volcano Plot\n(p-value vs Fold change)"), xlim =(-4,8))
ax.plot(X,y, marker='o',markersize=5, color = 'gray', linewidth=0)
# Add bordar line (log2(ratio)< -2, 2<log2(ratio))
ax.axvline(x= self.min_log2,color = 'black', linewidth = 0.5)
ax.axvline(x= self.max_log2, color = 'black', linewidth = 0.5)
# Add bordar line of p = 0.05
ax.axhline(y= p_cutoff, xmin=-4, xmax=8, color = 'black', linewidth = 0.5)
# Add text, p = 0.05
ax.text(8.1 , p_cutoff-0.05, 'p =' + str(self.pcutoff), size = 20, fontstyle="italic")
# Plot increasing genes
for j in range(0,len(gene_incraese)):
ax.plot(Xincrease[j], yincrease[j], marker='o', markersize=5, color = 'red', linewidth=0)
ax.annotate(gene_incraese[j],(Xincrease[j], yincrease[j]),
xytext=(0, 10), textcoords='offset points', size = 20, color = "black")
# Plot decreasing genes
for k in range(0,len(gene_decraese)):
ax.plot(Xdecrease[k], ydecrease[k], marker='o', markersize=5, color = 'blue', linewidth=0)
ax.annotate(gene_decraese[k],(Xdecrease[k], ydecrease[k]),
xytext=(0, 10), textcoords='offset points', size = 20, color = "black")
plt.show()
return df_increase, df_decrease
# set parameter and execute plot
p_cutoff = 0.05
min_log2ratio = -1
max_log2ratio = 1
volcano_plot = Volcano(df['ProteinID'],df['log2(ratio)'],df['(-log10(pvalue))'], p_cutoff,min_log2ratio,max_log2ratio)
inc,dec = volcano_plot.get_volcano()
# checking increased proteins
print(inc)
# checking decreased proteins
print(dec)
この記事が気に入ったらサポートをしてみませんか?
